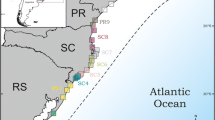
Abstract
Known for its remarkable biodiversity and high levels of endemism, the Brazilian Atlantic Rainforest has been characterized as one of the most threatened biomes on the planet. Despite strong interest in recent years, we still lack a comprehensive scenario to explain the origin and maintenance of diversity in this region, partially given the relatively low power of analyses involving few independent genetic loci. In this study, we examine a phylogenomic dataset of five ant species to investigate phylogeographical patterns across the Brazilian Atlantic Forest. We sequenced ultraconserved elements to generate hundreds of loci using a bait set developed specifically for hymenopterans. We analyzed the data using Bayesian and maximum likelihood approaches of phylogenetic inference. Results were then integrated with environmental niche modeling of current and past climates, including the Last Glacial Maximum and the last interglacial period. The studied species showed differentiation patterns that were consistent with the north/south division of the Atlantic Rainforest indicated in previous studies for other taxa. However, there were differences among species, both in the location of phylogeographic breaks and in the pattern of genetic variation within these areas. Samples from southern localities tended to show recent genetic structure, although a site in Tapiraí (state of São Paulo) repeatedly showed an intriguing older history of differentiation. All species experienced shifts in areas of suitability through the time. Our study suggests that distinct groups may have responded idiosyncratically to the climatic changes that took place in the Brazilian Atlantic Forest. The amount of intraspecific genetic structure was related to the inferred geographical distribution of habitat suitability according to current and past times. Also, a parallel between the amount of Quaternary climatic suitability and the level of interspecific differentiation was detected for four species. Finally, despite strong contractions at the northeastern region of the forest, the remaining areas appear to have been able to act as refugia.



Similar content being viewed by others
References
Aiello-Lammens, M., Boria, R., Radosavljevic, A., Vilela, B., & Anderson, R. (2015). spThin: an R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography, 38, 541–545. https://doi.org/10.1111/ecog.01132.
Álvarez-Presas, M., Sánchez-Gracia, A., Carbayo, F., Rozas, J., & Riutort, M. (2014). Insights into the origin and distribution of biodiversity in the Brazilian Atlantic Forest hot spot: a statistical phylogeographic study using a low-dispersal organism. Heredity, 112, 656–665. https://doi.org/10.1038/hdy.2014.3.
Amaro, R., Rodrigues, M., Yonenaga-Yassuda, Y., & Carnaval, A. (2012). Demographic processes in the montane Atlantic Rainforest: molecular and cytogenetic evidence from the endemic frog Proceratophrys boiei. Molecular Phylogenetics and Evolution, 62, 880–888. https://doi.org/10.1016/j.ympev.2011.11.004.
Andrews, S. (2010). FastQC—a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Baer, B. (2011). The copulation biology of ants (Hymenoptera: Formicidae). Myrmecological News, 14, 55–58.
Barve, N., & Barve, V. (2016). ENMGadgets: tools for pre and post processing in ENM workflow. R package version 0.0.12. https://github.com/narayanibarve/ENMGadgets. (accessed August 15, 2017).
Barve, N., Barve, V., Jiménez-Valverde, A., Lira-Noriega, A., Maher, S., Peterson, A., Soberón, J., & Villalobos, F. (2011). The crucial role of the accessible area in ecological niche modeling and species distribution modeling. Ecological Modelling, 222, 1810–1819. https://doi.org/10.1016/j.ecolmodel.2011.02.011.
Batalha-Filho, H., & Miyaki, C. (2016). Late Pleistocene divergence and postglacial expansion in the Brazilian Atlantic Forest: multilocus phylogeography of Rhopias gularis (Aves: Passeriformes). Journal of Zoological Systematics and Evolutionary Research, 54, 137–147. https://doi.org/10.1111/jzs.12118.
Batalha-Filho, H., Cabanne, G., & Miyaki, C. (2012). Phylogeography of an Atlantic Forest passerine reveals demographic stability through the Last Glacial Maximum. Molecular Phylogenetics and Evolution, 65, 892–902. https://doi.org/10.1016/j.ympev.2012.08.010.
Bernatchez, L., & Wilson, C. (1998). Comparative phylogeography of Nearctic and Palearctic fishes. Molecular Ecology, 7, 431–452. https://doi.org/10.1046/j.1365-294x.1998.00319.x.
Blaimer, B., Brady, S., Schultz, T., Lloyd, M., Fisher, B., & Ward, P. (2015). Phylogenomic methods outperform traditional multi-locus approaches in resolving deep evolutionary history: a case study of formicine ants. BMC Evolutionary Biology, 15, 271. https://doi.org/10.1186/s12862-015-0552-5.
Bolton, B. 2019. An online catalog of the ants of the world. Available from http://antcat.org. (accessed March 14, 2019).
Bonaccorso, E., Koch, I., & Townsend, P. A. (2006). Pleistocene fragmentation of Amazon species’ ranges. Diversity and Distributions, 12, 157–164. https://doi.org/10.1111/j.1366-9516.2005.00212.x.
Bragagnolo, C., Pinto-da-Rocha, R., Antunes, M., & Clouse, R. (2015). Phylogenetics and phylogeography of a long-legged harvestman (Arachnida: Opiliones) in the Brazilian Atlantic Rain Forest reveals poor dispersal, low diversity and extensive mitochondrial introgression. Invertebrate Systematics, 29, 386. https://doi.org/10.1071/IS15009.
Brown, W. L., Jr.; Kempf, W. W. (1960). A world revision of the ant tribe Basicerotini. Studia Entomologica (n.s.)3:161–250.
Cabanne, G., Calderón, L., Trujillo, A. N., Flores, P., Pessoa, R., d’Horta, F., & Miyaki, C. (2016). Effects of Pleistocene climate changes on species ranges and evolutionary processes in the Neotropical Atlantic Forest. Biological Journal of the Linnean Society, 119, 856–872. https://doi.org/10.1111/bij.12844.
Cardoso, D., Cristiano, M., Tavares, M., Schubart, C., & Heinze, J. (2015). Phylogeography of the sand dune ant Mycetophylax simplex along the Brazilian Atlantic Forest coast: remarkably low mtDNA diversity and shallow population structure. BMC Evolutionary Biology, 15, 106. https://doi.org/10.1186/s12862-015-0383-4.
Carnaval, A., & Moritz, C. (2008). Historical climate modelling predicts patterns of current biodiversity in the Brazilian Atlantic Forest. Journal of Biogeography, 35, 1187–1201. https://doi.org/10.1111/j.1365-2699.2007.01870.x.
Carnaval, A., Hickerson, M., Haddad, C., Rodrigues, M., & Moritz, C. (2009). Stability predicts genetic diversity in the Brazilian Atlantic Forest hotspot. Science, 323, 785–789. https://doi.org/10.1126/science.1166955.
Carnaval, A., Waltari, E., Rodrigues, M., Rosauer, D., VanDerWal, J., Damasceno, R., et al. (2014). Prediction of phylogeographic endemism in an environmentally complex biome. Proceedings of the Royal Society B: Biological Sciences, 281, 20141461–20141461. https://doi.org/10.1098/rspb.2014.1461.
Costa, L. (2003). The historical bridge between the Amazon and the Atlantic Forest of Brazil: a study of molecular phylogeography with small mammals. Journal of Biogeography, 30, 71–86. https://doi.org/10.1046/j.1365-2699.2003.00792.x.
Costa, L., Leite, Y., da Fonseca, G., & da Fonseca, M. (2000). Biogeography of South American forest mammals: endemism and diversity in the Atlantic Forest. Biotropica, 32, 872–881. https://doi.org/10.1111/j.1744-7429.2000.tb00625.x.
Cristiano, M. P., Cardoso, C. D., Fernandes-Salomão, T. M., & Heinze, J. (2016). Integrating paleodistribution models and phylogeography in the grass-cutting ant Acromyrmex striatus (Hymenoptera: Formicidae) in southern lowlands of South America. PLoS One, 11(1), e0146734. https://doi.org/10.1371/journal.pone.0146734.
Darwin, C., & Keynes, R. (2004). Charles Darwin’s “Beagle” diary. Cambridge: Cambridge University Press.
Drummond, A., Suchard, M., Xie, D., & Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution, 29, 1969–1973. https://doi.org/10.1093/molbev/mss075.
Elith, J., Graham, H. C., Anderson, P. R., Dudík, M., Ferrier, S., Guisan, A., et al. (2006). Novel methods improve prediction of species’ distributions from occurrence data. Ecography, 29, 129–151. https://doi.org/10.1111/j.2006.0906-7590.04596.x.
Faircloth, B. (2013). Illumiprocessor: a trimmomatic wrapper for parallel adapter and quality trimming. https://doi.org/10.6079/J9ILL; https://illumiprocessor.readthedocs.io/en/latest/citing.html. (accessed April 16, 2015).
Faircloth, B. (2015). PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics, 32, 786–788. https://doi.org/10.1093/bioinformatics/btv646.
Faircloth, B., & Glenn, T. (2012). Not all sequence tags are created equal: designing and validating sequence identification tags robust to indels. PLoS One, 7, e42543. https://doi.org/10.1371/journal.pone.0042543.
Faircloth, B., Branstetter, M., White, N., & Brady, S. (2015). Target enrichment of ultraconserved elements from arthropods provides a genomic perspective on relationships among Hymenoptera. Molecular Ecology Resources, 15, 489–501. https://doi.org/10.1111/1755-0998.12328.
Fick, S., & Hijmans, R. (2017). WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. International Journal of Climatology, 37, 4302–4315. https://doi.org/10.1002/joc.5086.
Fisher, S., Barry, A., Abreu, J., Minie, B., Nolan, J., Delorey, T., Young, G., et al. (2011). A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biology, 12, R1. https://doi.org/10.1186/gb-2011-12-1-r1.
Forel, A. (1905). Miscellanea myrmécologiques II (1905). Annales de la Société Entomologique de Belgique 49:155–185.
Garrick, R., Sands, C., Rowell, D., Tait, N., Greenslade, P., & Sunnucks, P. (2004). Phylogeography recapitulates topography: very fine-scale local endemism of a saproxylic ‘giant’ springtail at Tallaganda in the great dividing range of south-east Australia. Molecular Ecology, 13, 3329–3344. https://doi.org/10.1111/j.1365-294X.2004.02340.x.
Giraudo, A., Matteucci, S., Alonso, J., Herrera, J., & Abramson, R. (2008). Comparing bird assemblages in large and small fragments of the Atlantic Forest hotspots. Biodiversity and Conservation, 17, 1251–1265. https://doi.org/10.1007/s10531-007-9309-9.
Glenn, T., Nilsen, R., Kieran, T., Finger, J., Pierson, T., Bentley, K., et al. (2016). Adapterama I: universal stubs and primers for thousands of dual-indexed Illumina libraries (iTru and iNext). bioRxiv. https://doi.org/10.1101/049114.
Gnirke, A., Melnikov, A., Maguire, J., Rogov, P., LeProust, E., Brockman, W., et al. (2009). Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nature Biotechnology, 27, 182–189. https://doi.org/10.1038/nbt.1523.
Grabherr, M., Haas, B., Yassour, M., Levin, J., Thompson, D., Amit, I., & Regev, A. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology, 29, 644–652. https://doi.org/10.1038/nbt.1883.
Grazziotin, F., Monzel, M., Echeverrigaray, S., & Bonatto, S. (2006). Phylogeography of the Bothrops jararaca complex (Serpentes: Viperidae): past fragmentation and island colonization in the Brazilian Atlantic Forest. Molecular Ecology, 15, 3969–3982. https://doi.org/10.1111/j.1365-294X.2006.03057.x.
Haffer, J. (1969). Speciation in amazonian forest birds. Science, 165, 131–137. https://doi.org/10.1126/science.165.3889.131.
Haffer, J. (1997). Alternative models of vertebrate speciation in Amazonia: an overview. Biodiversity and Conservation, 6, 451–476. https://doi.org/10.1023/A:1018320925954.
Hare, M. (2001). Prospects for nuclear gene phylogeography. Trends in Ecology and Evolution, 16, 700–706. https://doi.org/10.1016/S0169-5347(01)02326-6.
Hayes, F., & Sewlal, J. (2004). The Amazon River as a dispersal barrier to passerine birds: effects of river width, habitat and taxonomy. Journal of Biogeography, 31, 1809–1818. https://doi.org/10.1111/j.1365-2699.2004.01139.x.
Hijmans, R., Cameron, S., Parra, J., Jones, P., & Jarvis, A. (2005). Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology, 25, 1965–1978.
Hölldobler, B., & Wilson, E. (1990). The ants. Cambridge: Belknap Press of Harvard University Press.
Katoh, K., & Standley, D. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780. https://doi.org/10.1093/molbev/mst010.
Lara, M., & Patton, J. (2000). Evolutionary diversification of spiny rats (genus Trinomys, Rodentia: Echimyidae) in the Atlantic Forest of Brazil. Zoological Journal of the Linnean Society, 130, 661–686. https://doi.org/10.1111/j.1096-3642.2000.tb02205.x.
Leão, T., Fonseca, C., Peres, C., & Tabarelli, M. (2014). Predicting extinction risk of Brazilian Atlantic Forest angiosperms. Conservation Biology, 28, 1349–1359. https://doi.org/10.1111/cobi.12286.
Leite, Y., Costa, L., Loss, A., Rocha, R., Batalha-Filho, H., Bastos, A., et al. (2016a). Neotropical forest expansion during the last glacial period challenges refuge hypothesis. Proceedings of the National Academy of Sciences, 113, 1008–1013. https://doi.org/10.1073/pnas.1513062113.
Leite, Y., Costa, L., Loss, A., Rocha, R., Batalha-Filho, H., Bastos, A., et al. (2016b). Reply to Raposo do Amaral et al.: The “Atlantis Forest hypothesis” adds a new dimension to Atlantic Forest biogeography. Proceedings of the National Academy of Sciences, 113, E2099–E2100. https://doi.org/10.1073/pnas.1602391113.
Leppänen, J., Vepsäläinen, K., & Savolainen, R. (2011). Phylogeography of the ant Myrmica rubra and its inquiline social parasite. Ecology and Evolution, 1, 46–62. https://doi.org/10.1002/ece3.6.
Lohse, M., Bolger, A., Nagel, A., Fernie, A., Lunn, J., Stitt, M., & Usadel, B. (2012). RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Research, 40, W622–W627. https://doi.org/10.1093/nar/gks540.
Maddison, W., Knowles, L., & Collins, T. (2006). Inferring phylogeny despite incomplete lineage sorting. Systematic Biology, 55, 21–30. https://doi.org/10.1080/10635150500354928.
Martins, F., Templeton, A., Pavan, A., Kohlbach, B., & Morgante, J. (2009). Phylogeography of the common vampire bat (Desmodus rotundus): arked population structure, neotropical pleistocene vicariance and incongruence between nuclear and mtDNA markers. BMC Evolutionary Biology, 9, 294. https://doi.org/10.1186/1471-2148-9-294.
Mayr, G. 1887. Südamerikanische Formiciden. Verhandlungen der Kaiserlich-Königlichen Zoologisch-Botanischen Gesellschaft in Wien 37:511–632
Merow, C., Smith, M., & Silander, J. (2013). A practical guide to MaxEnt for modeling species’ distributions: what it does, and why inputs and settings matter. Ecography, 36, 1058–1069. https://doi.org/10.1111/j.1600-0587.2013.07872.x.
Morales, N., Fernández, I., Carrasco, B., & Orchard, C. (2015). Combining niche modelling, land-use change, and genetic information to assess the conservation status of Pouteria splendens populations in Central Chile. International Journal of Ecology, 2015, 1–12. https://doi.org/10.1155/2015/612194.
Murray-Smith, C., Brummitt, N., Oliveira-Filho, A., Bachman, S., Moat, J., Lughadha, E., & Lucas, E. (2009). Plant diversity hotspots in the Atlantic coastal forests of Brazil. Conservation Biology, 23, 151–163. https://doi.org/10.1111/j.1523-1739.2008.01075.x.
Myers, N., Mittermeier, R., Mittermeier, C., da Fonseca, G., & Kent, J. (2000). Biodiversity hotspots for conservation priorities. Nature, 403, 853–858. https://doi.org/10.1038/35002501.
Nicolas, V., Martínez-Vargas, J., & Hugot, J. (2016). Molecular data and ecological niche modelling reveal the evolutionary history of the common and Iberian moles (Talpidae) in Europe. Zoologica Scripta, 46, 12–26. https://doi.org/10.1111/zsc.12189.
Oliveira, U., Vasconcelos, M., & Santos, A. (2017). Biogeography of Amazon birds: rivers limit species composition, but not areas of endemism. Scientific Reports, 7, 2992. https://doi.org/10.1038/s41598-017-03098-w.
Pearson, R., Raxworthy, C., Nakamura, M., & Townsend, P. A. (2006). Predicting species distributions from small numbers of occurrence records: a test case using cryptic geckos in Madagascar. Journal of Biogeography, 34, 102–117. https://doi.org/10.1111/j.1365-2699.2006.01594.x.
Pellegrino, K., Rodrigues, M., Waite, A., Morando, M., Yassuda, Y., & Sites, J. (2005). Phylogeography and species limits in the Gymnodactylus darwinii complex (Gekkonidae, Squamata): genetic structure coincides with river systems in the Brazilian Atlantic Forest. Biological Journal of the Linnean Society, 85, 13–26. https://doi.org/10.1111/j.1095-8312.2005.00472.x.
Peres, E. A., Sobral-Souza, T., Perez, M. F., Bonatelli, I. A. S., Silva, D. P., Silva, M. J., & Solferini, V. N. (2015). Pleistocene niche stability and lineage diversification in the subtropical spider Araneus omnicolor (Araneidae). PLoS One, 10(4), e0121543. https://doi.org/10.1371/journal.pone.0121543.
Peterson, A., Soberón, J., Pearson, R., Anderson, R., Nakamura, M., Martinez-Meyer, E., & Araújo, M. (2011). Ecological niches and geographical distributions. Princeton: Princeton University Press.
Phillips, S., Anderson, R., Dudík, M., Schapire, R., & Blair, M. (2017). Opening the black box: an open-source release of Maxent. Ecography, 40, 887–893. https://doi.org/10.1111/ecog.03049.
Prates, I., Xue, A., Brown, J., Alvarado-Serrano, D., Rodrigues, M., Hickerson, M., & Carnaval, A. (2016). Inferring responses to climate dynamics from historical demography in neotropical forest lizards. Proceedings of the National Academy of Sciences, 113, 7978–7985. https://doi.org/10.1073/pnas.1601063113.
Quek, S., Davies, S., Ashton, P., Itino, T., & Pierce, N. (2007). The geography of diversification in mutualistic ants: a gene’s-eye view into the neogene history of Sundaland rain forests. Molecular Ecology, 16, 2045–2062. https://doi.org/10.1111/j.1365-294X.2007.03294.x.
Raposo do Amaral, F., Edwards, S., Pie, M., Jennings, W., Svensson-Coelho, M., d’Horta, F., Schmitt, C., & Maldonado-Coelho, M. (2016). The “Atlantis Forest hypothesis” does not explain Atlantic Forest phylogeography. Proceedings of the National Academy of Sciences, 113, E2097–E2098. https://doi.org/10.1073/pnas.1602213113.
Resende, H., Yotoko, K., Delabie, J., Costa, M., Campiolo, S., Tavares, M., Campos, L., & Fernandes-Salomão, T. (2010). Pliocene and Pleistocene events shaping the genetic diversity within the central corridor of the Brazilian Atlantic Forest. Biological Journal of the Linnean Society, 101, 949–960. https://doi.org/10.1111/j.1095-8312.2010.01534.x.
Ribeiro, M., Metzger, J., Martensen, A., Ponzoni, F., & Hirota, M. (2009). The Brazilian Atlantic Forest: how much is left, and how is the remaining forest distributed? Implications for conservation. Biological Conservation, 142, 1141–1153. https://doi.org/10.1016/j.biocon.2009.02.021.
Ribeiro, R., Lemos-Filho, J., Ramos, A., & Lovato, M. (2010). Phylogeography of the endangered rosewood Dalbergia nigra (Fabaceae): insights into the evolutionary history and conservation of the Brazilian Atlantic Forest. Heredity, 106, 46–57. https://doi.org/10.1038/hdy.2010.64.
Rohland, N., & Reich, D. (2012). Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Research, 22, 939–946. https://doi.org/10.1101/gr.128124.111.
Seal, J., Brown, L., Ontiveros, C., Thiebaud, J., & Mueller, U. (2015). Gone to Texas: phylogeography of two Trachymyrmex (Hymenoptera: Formicidae) species along the southeastern coastal plain of North America. Biological Journal of the Linnean Society, 114, 689–698. https://doi.org/10.1111/bij.12426.
Silva, J., & Casteleti, C. (2003). Status of the biodiversity of the Atlantic Forest of Brazil. The Atlantic Forest of South America: biodiversity status, threats, and outlook. (ed. by C. Galindo-Leal and I. Câmara), pp. 43–59. CABS and Island Press, Washington.
Silva, J., Cardoso de Sousa, M., & Castelletti, C. (2004). Areas of endemism for passerine birds in the Atlantic Forest, South America. Global Ecology and Biogeography, 13, 85–92. https://doi.org/10.1111/j.1466-882X.2004.00077.x.
Silva, J., Rylands, A., & da Fonseca, G. (2005). The fate of the amazonian areas of endemism. Conservation Biology, 19, 689–694. https://doi.org/10.1111/j.1523-1739.2005.00705.x.
Simpson, B. (1979). Quaternary biogeography of the high montane regions of South America. The south American Herpetofauna: its origin, evolution, and dispersal (ed. by W. Duellman), pp. 157–188. Monograph of the Museum of Natural History, University of Kansas.
Smith, B., Harvey, M., Faircloth, B., Glenn, T., & Brumfield, R. (2013). Target capture and massively parallel sequencing of ultraconserved elements for comparative studies at shallow evolutionary time scales. Systematic Biology, 63, 83–95. https://doi.org/10.1093/sysbio/syt061.
Smith, B., McCormack, J., Cuervo, A., Hickerson, M., Aleixo, A., Cadena, C., et al. (2014). The drivers of tropical speciation. Nature, 515, 406–409. https://doi.org/10.1038/nature13687.
Solomon, S., Bacci, M., Martins, J., Vinha, G., & Mueller, U. (2008). Paleodistributions and comparative molecular phylogeography of leafcutter ants (Atta spp.) provide new insight into the origins of amazonian diversity. PLoS One, 3, e2738. https://doi.org/10.1371/journal.pone.0002738.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312–1313. https://doi.org/10.1093/bioinformatics/btu033.
Ströher, P. R., Li, C., & Pie, M. (2013). Exon-primed intron-crossing (EPIC) markers as a tool for ant phylogeography. Revista Brasileira de Entomologia, 57, 427–430. https://doi.org/10.1590/S0085-56262013005000039.
Tchaicka, L., Eizirik, E., De Oliveira, T., Cândido, J., & Freitas, T. (2006). Phylogeography and population history of the crab-eating fox (Cerdocyon thous). Molecular Ecology, 16, 819–838. https://doi.org/10.1111/j.1365-294X.2006.03185.x.
Thomé, M., Zamudio, K., Giovanelli, J., Haddad, C., Baldissera, F., & Alexandrino, J. (2010). Phylogeography of endemic toads and post-Pliocene persistence of the Brazilian Atlantic Forest. Molecular Phylogenetics and Evolution, 55, 1018–1031. https://doi.org/10.1016/j.ympev.2010.02.003.
Thomé, M., Zamudio, K., Haddad, C., & Alexandrino, J. (2014). Barriers, rather than refugia, underlie the origin of diversity in toads endemic to the Brazilian Atlantic Forest. Molecular Ecology, 23, 6152–6164. https://doi.org/10.1111/mec.12986.
Vanzolini, P. (1992). Paleoclimas e especiação em animais da América do Sul tropical. Estudos Avançados, 6, 41–65. https://doi.org/10.1590/S0103-40141992000200003.
Vanzolini, P., & Williams, E. (1981). Vanishing refuge: a mechanism for ecogeographic speciation. Papéis Avulsos de Zoologia, 34, 251–255.
Wenger, S., & Olden, J. (2012). Assessing transferability of ecological models: an underappreciated aspect of statistical validation. Methods in Ecology and Evolution, 3, 260–267. https://doi.org/10.1111/j.2041-210X.2011.00170.x.
Zamudio, K., & Greene, H. (1997). Phylogeography of the bushmaster (Lachesis muta: Viperidae): implications for neotropical biogeography, systematics, and conservation. Biological Journal of the Linnean Society, 62, 421–442. https://doi.org/10.1111/j.1095-8312.1997.tb01634.x.
Zwiener, V., Padial, A., Marques, M., Faleiro, F., Loyola, R., & Peterson, A. (2017). Planning for conservation and restoration under climate and land use change in the Brazilian Atlantic Forest. Diversity and Distributions, 23, 955–966. https://doi.org/10.1111/ddi.12588.
Acknowledgements
We thank Rogerio R. Silva for providing the specimens used in this project and Rodrigo M. Feitosa for the taxonomic advice. We are grateful to the two anonymous reviewers for their helpful comments that greatly improved the manuscript. We also would like to thank Eduardo A. B. de Almeida for the international and financial logistics support.
Funding
This study was funded by a grant from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (301636/2016-8) to MRP. This study was also funded by scholarships from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) to Patrícia Regina Ströher (Processo: PDSE 99999.002880/2014-08) and from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (140262/2013-0).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 15 kb)
Rights and permissions
About this article
Cite this article
Ströher, P.R., Meyer, A.L.S., Zarza, E. et al. Phylogeography of ants from the Brazilian Atlantic Forest. Org Divers Evol 19, 435–445 (2019). https://doi.org/10.1007/s13127-019-00409-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-019-00409-z